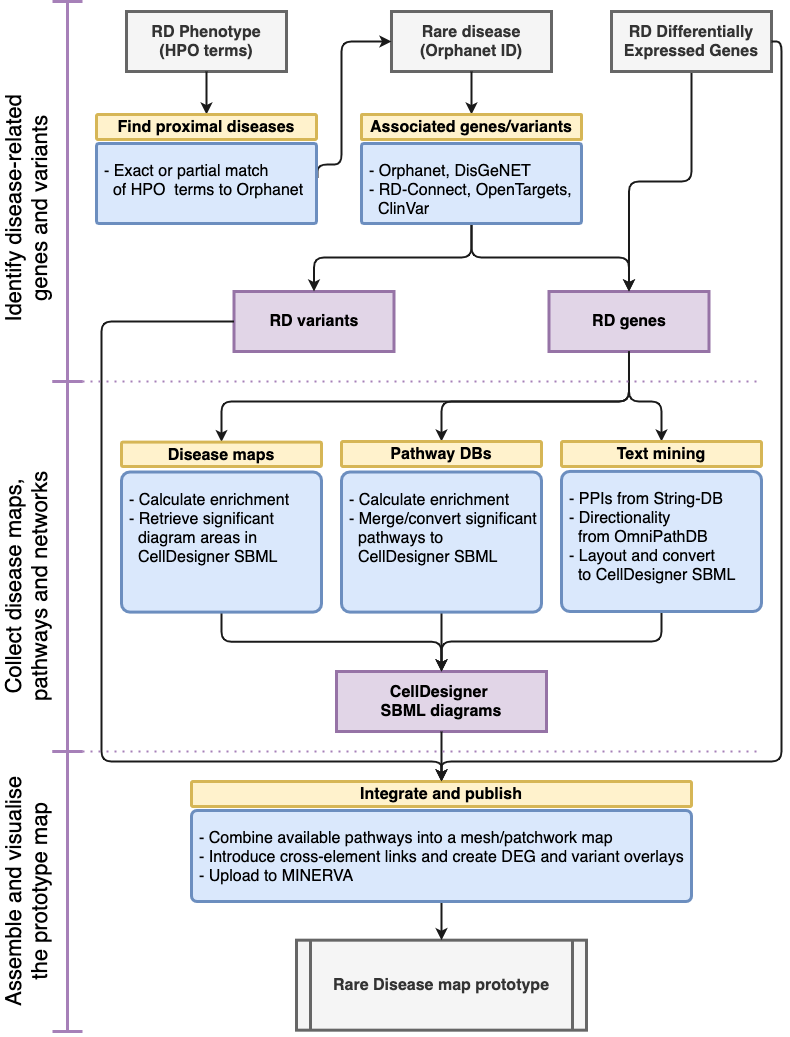
Goal
To make it easier to build disease maps (see Disease Maps Community website>), as this is a time-consuming task. Disease maps support research on molecular mechanisms, but their development requires dedicated resources. we want so specifically help Rare Diseases (RD), as these disorders are not sufficiently represented across major bioinformatics resources or pathway databases. We want to systematically scan existing disease-focused resources and offer insights into specific RDs.